Downloading Publicly Available Data
Data submitted to NeMO Archive falls into three categories:
- Public - data to be immediately distributed openly and freely to the wider research community,
- Embargo - data to be held back, or embargoed, until a specific date, at which point it will be released openly and freely to the wider research community,
- Restricted - Controlled access data to be distributed only to an approved group of users due to consent restrictions, e.g. human data. Often restricted datasets contain a combination of private (raw reads, alignments) and public (counts, peaks) datatypes. In such instances, the dataset landing page or BDBag will provide direct access to public data, in addition to a link to restricted data access instructions.
While we endeavor to make accessing data on NeMO Archive as quick as possible, in cases of extraordinary activity (e.g. downloading all public data present in the archive) we may temporarily limit the bandwith available to individual users to ensure activites do not cause undue slowdowns in site performance.
There are many ways that users may be introduced to data at NeMO Archive, depending on whether you're arriving at our site directly or from another resource. For Restricted data access, see the documentation here. For public data users, we've put together this handy table to recommend the most appropriate way to access data:
| So you want to... | We recommend... |
|---|---|
| Access a specific dataset from a publication | Collection landing pages |
| Access a specific data collection found in the BICCN Data Inventory | Collection landing pages |
| Browse to explore the types of data available across the project | HTTP Browser |
| Download an entire NeMO release by date | HTTP Browser |
| Download all data associated with a particular grant | HTTP Browser or Data Portal |
| Download all data generated by a particular lab | HTTP Browser or Data Portal |
| Download all data associated with a particular technique | Data Portal |
| Download all data from a specific species (mouse, marmoset, macaque) | Data Portal |
| Download all counts data | Data Portal |
| Download all data from a particular anatomical site | Data Portal |
| Pull data into NeMO Analytics, Terra, Metaviz and other tools to run custom pipelines | Data Portal |
Each of these access points is introduced below, with links to further documentation if necessary.
Data collection landing pages
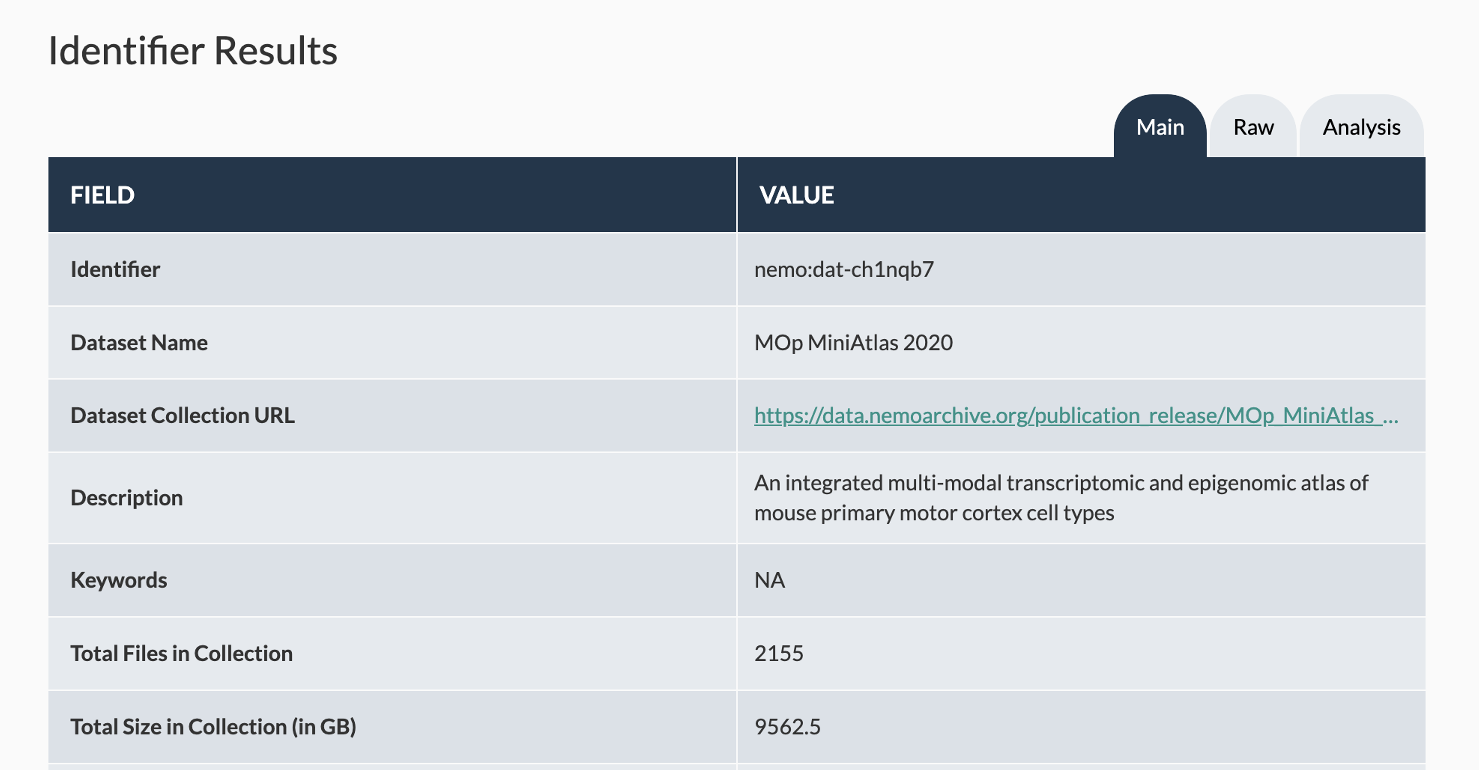
Landing pages are available at assets.nemoarchive.org. We are integrating a search function to allow users to search by NeMO identifier, but in the meantime, you can pull up any dataset of interest by adding the NeMO identifier to the end of the url, e.g.
assets.nemoarchive.org/dat-ch1nqb7Each data collection landing page corresponds to a defined dataset, either from a publication or a direct link from biccn.org. Landing pages contain basic metadata about the dataset, along with links to either the HTTP browser (see next section) if the dataset corresponds to a single branch of the directory structure, or a BDBag if it contains a subset of data or data across multiple modalities, labs or cell types. More information about downloading and working with BDBags is available here.
If you are a PI or a data submitter and would like to organize a data collection into a BDBag and/or request a landing page for a data collection or publication, please contact the NeMO Archive team.
HTTP directory structure browser
An HTTP server-based browser is available at data.nemoarchive.org for navigation through the NeMO Archive public data directory structure. Data can be downloaded from the NeMO HTTP browser using any tools that support http downloads.
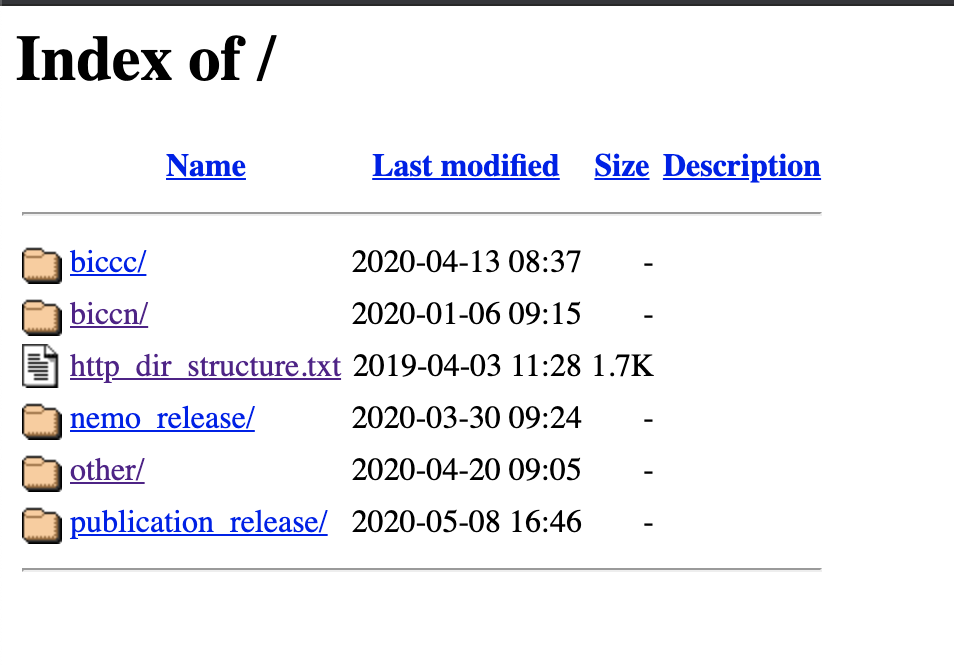
The top, or root, level of the HTTP browser separates data based on release or project:
Releases
nemo_release/contains a BDBag corresponding to quarterly NeMO releases. NeMO releases are synchronized to the same release schedule as the BCDC release schedule. These bags contain BICCN data only.publication_release/contains one or more BDBags corresponding to dataset(s) analyzed for BICCN-associated publications. See Data citation for more information.
Projects
biccc/contains data generated within the BRAIN Initiative Cell Census Consortium, precursor to the BICCNbiccn/contains data generated as part of the ongoing BRAIN Initiative Cell Census Networkother/In addition to hosting BRAIN Initiative data, the NeMO repository also hosts 'omics data from other neuroscience projects. Contact us if you would like to discuss submission of your dataset(s) to the NeMO Archives.
Within each project, data is organized by grant, lab, organism or modality. Read more about the data storage model.
NeMO Data Portal
The NeMO Data Portal provides faceted search and advanced query tools to enable users to explore data in a more customized way. To help users find data, the NeMO Data Portal uses a structured query approach. The interface offers a simple faceted search mode along with an advanced search mode. The faceted search uses a predefined subset of curated popular metadata fields as facets, including grants, organism, anatomical structures, data modality, library methods, and file format. Through a simple point-and-click interface, the user can generate a summary of the number of samples, file types and data sizes matching their selected criteria. The summary is displayed graphically as a set of interactive pie charts that can be clicked on to further refine the search. Once a user has identified a set of files of interest, they can add this set to a data cart for further operations, including direct download or export to Terra.
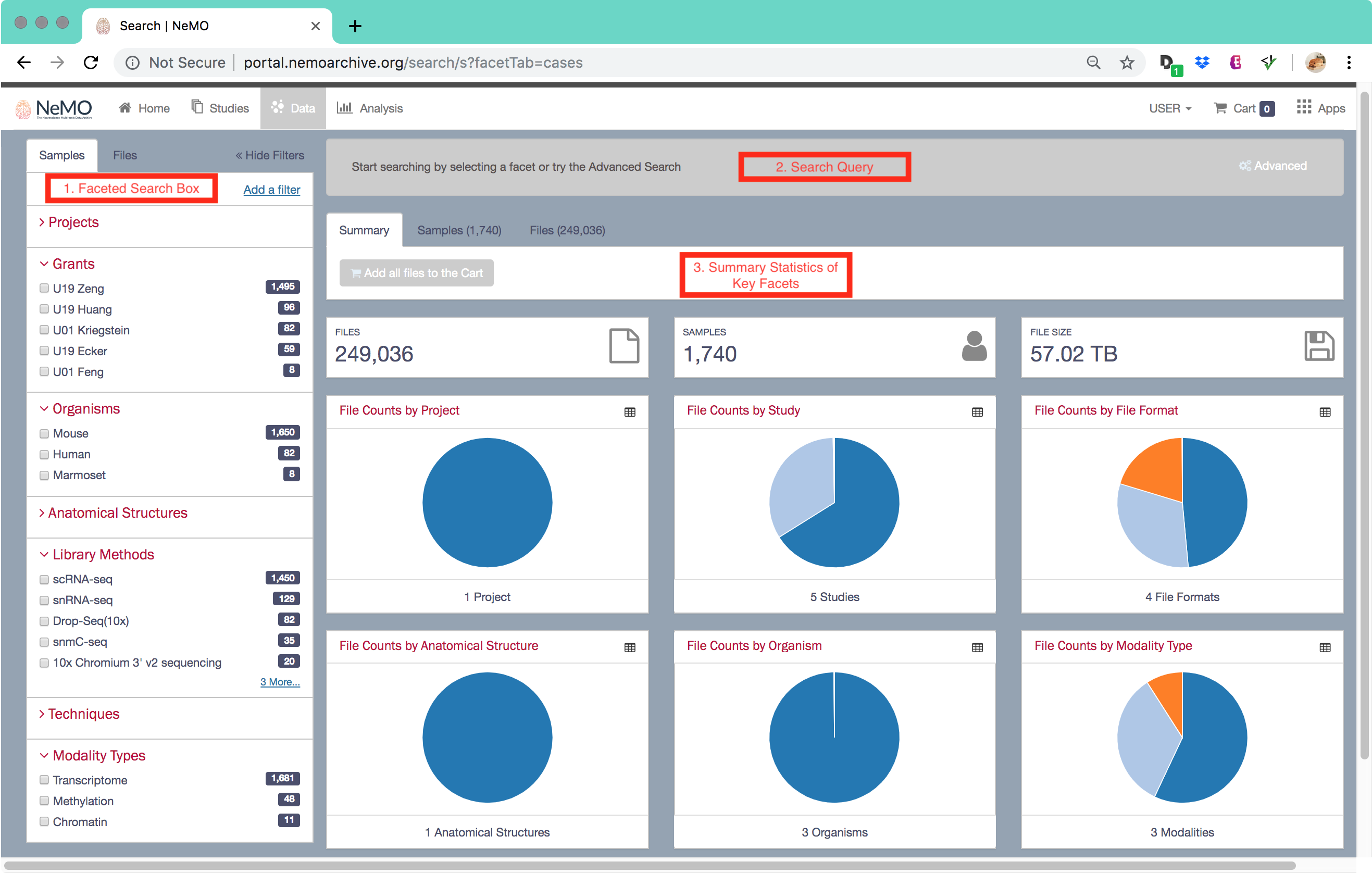
Additional NeMO Data Portal documentation can be found at the following pages: