Accessing Controlled Data
Data submitted to NeMO falls into three categories:
- Public - data to be immediately distributed openly and freely to the wider research community,
- Embargo - data to be held back, or embargoed, until a specific date, at which point it will be released openly and freely to the wider research community,
- Restricted - Controlled access data to be distributed only to an approved group of users due to consent restrictions, e.g. human data, through the processes described here. Often restricted datasets contain a combination of private (raw reads, alignments) and public (counts, peaks) datatypes. In such instances, the dataset landing page or BDBag will provide direct access to public data, in addition to a link to this page for restricted data access instructions.
You can see the current list of projects available for access request by logging into the NIMH Data Archive and selecting Get Data > Request Access. Under NDA Controlled Access permissions Groups, you will find all BRAIN/NeMO restricted datasets currently available for access request.
In this document:
- Restricted access datasets currently available for request at NDA
- Requesting access through the NDA Approval Process
- Restricted access datasets currently available for request at Broad
- Requesting access through the Broad's DAC Approval Process
- Downloading Data
Restricted access datasets currently available for request at NDA
The NIMH Data Archive will review data access requests in consideration of the data use limitations on these datasets before approval. For further instructions on how to request and get access to these datasets, go to Requesting access through the NDA Approval Process.
| Dataset | Description | Data Use Limitations | BICCN Webpage | Dataset Page |
|---|---|---|---|---|
| BRAIN/NeMO: A multimodal atlas of human brain cell types: Patch-Seq | This study (NIH Grant 1U01MH114812-01; PI: Lein, Ed) is part of an international consortium approach using single cell transcriptomics, human cellular physiology and anatomy and neuronal modeling to begin to create an atlas of human brain cell types. In this dataset, transcriptomic profiles of individual human cortical neurons were assayed by the SMART-seq v4 single nucleus RNA-seq method following acute brain slice patch-clamp electrophysiology recordings and nucleus extraction, i.e. the Patch-seq method. | Use of data is limited to brain in health and disease | Glutamatergic Neuron Types in Layer 2 and Layer 3 of the Human Cortex | dat-18e8800 |
| BRAIN/NeMO: Cell type-specific 3D epigenomes in the developing human cortex | This NIH grant U01DA052713 (PI: Shen, Yin) studies cis-regulatory chromatin interactions, open chromatin peaks, and transcriptomes for radial glia, intermediate progenitor cells, excitatory neurons, and interneurons isolated from mid-gestational human cortex samples. | These data have a data use limitation of General Research Use. Use of the data is limited only by the terms of the Data Use Certification and the requestor must provide documentation of local IRB approval. | dat-uioqy8b | |
| BRAIN/NeMO: A Cellular Resolution Census of the Developing Human Brain | This NIH grant U01MH114825 (PI: Kriegstein, Arnold) aims to create a spatiotemporal single cell resolution map of the developing human neocortex to establish how many distinct cell types are present and to unravel their complex developmental history. | Use of the data is limited to health, medical, biomedical purposes and use of the data is limited to not-for-profit organizations. | Single-Cell Sequencing of the Developing Human Brain | dat-t4xnmho (10x v2); dat-qf43d3z (10x v3) |
| BRAIN/NeMO: A Multimodal atlas of human brain cell types: Human variation RNAseq & WGS (Lein) | This dataset funded by Allen Institute for Brain Science (PI: Lein, Ed) is part of a multimodal investigation into human variation of brain cell types using RNA sequencing and whole genome shotgun sequencing. | Use of the data is limited to General Research Use (GRU). | Characterization of transcriptomic and genomic variation in adult human cortex | dat-3seoxdg (10x); dat-ofcr6j0 (wgs) |
| BRAIN/NeMO: A Multimodal atlas of human brain cell types: Human variation RNAseq & WGS, Limited to Brain Health & Disease (Lein) | This dataset is associated with permission group ‘BRAIN/NeMO: A Multimodal atlas of human brain cell types: Human variation RNAseq & WGS’. Access approval for this permission group WILL include access to the associated GRU permission group, please do not apply for access to both groups. | Use of this dataset is limited to Brain Health & Disease. | Characterization of transcriptomic and genomic variation in adult human cortex | dat-3seoxdg (10x); dat-ofcr6j0 (wgs) |
| BRAIN/NeMO: A multimodal atlas of human brain cell types: Human Cortex RNASeq | This study is part of an international consortium approach using single cell transcriptomics, human cellular physiology and anatomy and neuronal modeling to begin to create an atlas of human brain cell types. In this dataset, single nucleus RNA-sequencing using SMART-Seq v4 methodology was applied to perform a comprehensive analysis of cell types in a variety of human cortical areas (MTG, M1C, CgGr, V1C, S1C and A1C), largely from postmortem brain. | Limited to General Research Use (GRU) | Human Multiple Cortical Areas-SMART-seq; Cellular diversity in human, non-human primate, and mouse LGN; An atlas of human-specialized cortical cell types | dat-2c8otht; dat-9fbmh99 |
| BRAIN/NeMO: A multimodal atlas of human brain cell types: Middle Temporal Gyrus & Human Cortex RNASeq | This study is part of an international consortium approach using single cell transcriptomics, human cellular physiology and anatomy and neuronal modeling to begin to create an atlas of human brain cell types. In this dataset, single nucleus RNA-sequencing using SMART-Seq v4 methodology was applied to perform a comprehensive analysis of cell types from middle temporal gyrus (MTG) from neurosurgical tissue and in a variety of human cortical areas (MTG, M1C, CgGr, V1C, S1C and A1C), largely from postmortem brain. | Human cortical RNAseq data is limited to General Research Use (GRU) and the Middle Temporal Gyrus data is limited to Health/Medical/Biomedical (HMB). | Human Multiple Cortical Areas-SMART-seq | dat-swzf4kc |
| BRAIN/NeMO: Single Cell ATACseq of the Developing Human Brain (Nowakowski) | This dataset, funded under NIH grant 1U01MH114825-01 (PI: Kriegstein, Arnold) is part of an investigation into chromatin accessibility of cells from eight distinct areas of the developing human forebrain using single cell ATAC-seq (scATACseq). | Use of the data is limited to General Research Use (GRU). | Single-Cell ATAC-seq of the Developing Human Brain | dat-dgarbc3 |
| BRAIN/NeMO: Single-cell analysis of prenatal and postnatal human cortical development | Controlled access to genomics data housed at the BRAIN/NeMO data archive. This study (NIH grant U01MH114825, PI: Kriegstein, Arnold) aims to create a spatiotemporal single cell resolution map of the developing human neocortex to establish how many distinct cell types are present and to unravel their complex developmental history. These data have a consent-based data use limitation of General Research Use. The BRAIN/NeMO DAC will review data access requests in consideration of this data use limitation. For further instructions on how to access this dataset, go to https://nemoarchive.org/resources/accessing-controlled-access-data.php. | Use of the data is limited to General Research Use (GRU). | Single-Nuclei Sequencing of Late Stages of Human Brain Development and Early Postnatal Life | dat-b3brzfa |
| BRAIN/NeMO: A single-cell genomic atlas for maturation of the human cerebellum during early childhood | This dataset, funded under NIH grant R21MH128462 (PI: Seth Ament) is part of an investigation involving single-cell RNA sequencing of the post-mortem human cerebellum characterized transcriptional changes in Purkinje and Golgi neurons during early childhood. The study revealed that these developmental gene regulatory programs are prematurely down-regulated in the brains of children who perished under conditions that included inflammation compared to those who succumbed to sudden accidental death. These data have a consent-based data use limitation of General Research Use. The BRAIN/NeMO DAC will review data access requests in consideration of this data use limitation. For further instructions on how to access this dataset, go to https://nemoarchive.org/resources/accessing-controlled-access-data.php. | Use of the data is limited to General Research Use (GRU). | dat-sxfwwo8 | |
| BRAIN/NeMO: Developmental isoform diversity in the human neocortex informs neuropsychiatric risk mechanisms | This dataset, funded under NIH grant R01MH124018 (PI: Luis de la Torre-Ubieta) is part of an investigation involving single-molecule long-read sequencing to deeply profile the full-length transcriptome of the germinal zone (GZ) and cortical plate (CP) regions of the developing human neocortex at tissue and single-cell resolution to uncover contribution of transcript-isoform diversity, regulated by RNA binding proteins, in defining cellular identity in the developing neocortex and to inform genetic risk mechanisms for neuropsychiatric disorders. These data have a consent-based data use limitation of General Research Use. The BRAIN/NeMO DAC will review data access requests in consideration of this data use limitation. For further instructions on how to access this dataset, go to https://nemoarchive.org/resources/accessing-controlled-access-data.php. | Use of the data is limited to General Research Use (GRU) and not-for-profit organizations. The requestor must provide documentation of local IRB approval. | dat-rhocguc | |
| BRAIN/NeMO: Epigenomic and chromosomal architectural reconfiguration in developing human frontal cortex and hippocampus | This dataset, funded under NIH grant R01MH125252 (PI: Luo Chongyuan) is part of an investigation involving the epigenomic and 3D chromatin conformational reorganization during the development of the frontal cortex and hippocampus, using more than 53,000 joint single-nucleus profiles of chromatin conformation and DNA methylation (sn-m3C-seq). The remodeling of DNA methylation predominantly occurs during late-gestational to early-infant development and is temporally separated from chromatin conformation dynamics. Neurons have a unique Domain-Dominant chromatin conformation that is different from the Compartment-Dominant conformation of glial cells and non-brain tissues. The project involved reconstruction of the regulatory programs of cell-type differentiation and finding putatively causal common variants for schizophrenia that strongly overlapped with chromatin loop-connected, cell-type-specific regulatory regions. The data demonstrate that single-cell 3D-regulome is an effective approach for dissecting neuropsychiatric risk loci. These data have a consent-based data use limitation of General Research Use. The BRAIN/NeMO DAC will review data access requests in consideration of this data use limitation. For further instructions on how to access this dataset, go to https://nemoarchive.org/resources/accessing-controlled-access-data.php. | Use of the data is limited to General Research Use (GRU). | dat-obec38w | |
| BRAIN/NeMO: Single-Cell Multi-Omic Analysis of the Developing Human Brain | This dataset is funded under 1U01MH114825 (PI: Kriegstein, Arnold). To understand the genetic and epigenetic programs that control human cortex development, we simultaneously profiled the gene expression and chromatin accessibility from the same cell using the 10x Single-Cell Multiome assay in multiple human cerebral cortical areas from the first trimester to the late childhood stage. Areas analyzed include the prefrontal cortex and the primary visual cortex. These data could be used to infer the gene regulatory networks that control cellular differentiation in individual cell types and lineages. These data have a consent-based data use limitation of General Research Use. The BRAIN/NeMO DAC will review data access requests in consideration of this data use limitation. For further instructions on how to access this dataset, go to https://nemoarchive.org/resources/accessing-controlled-access-data.php. | Use of the data is limited to General Research Use (GRU). | Single-Cell Multi-Omic Analysis of the Developing Human Brain | dat-oiif74w |
NDA Approval Process
Permissions for restricted data access at NeMO are being facilitated by the NIMH Data Archive (NDA). NDA and NeMO are working together to ensure a smooth process. NDA provides an SOP for institutionally sponsored data access requests here, however this page outlines the steps required for NeMO-specific restricted data access.

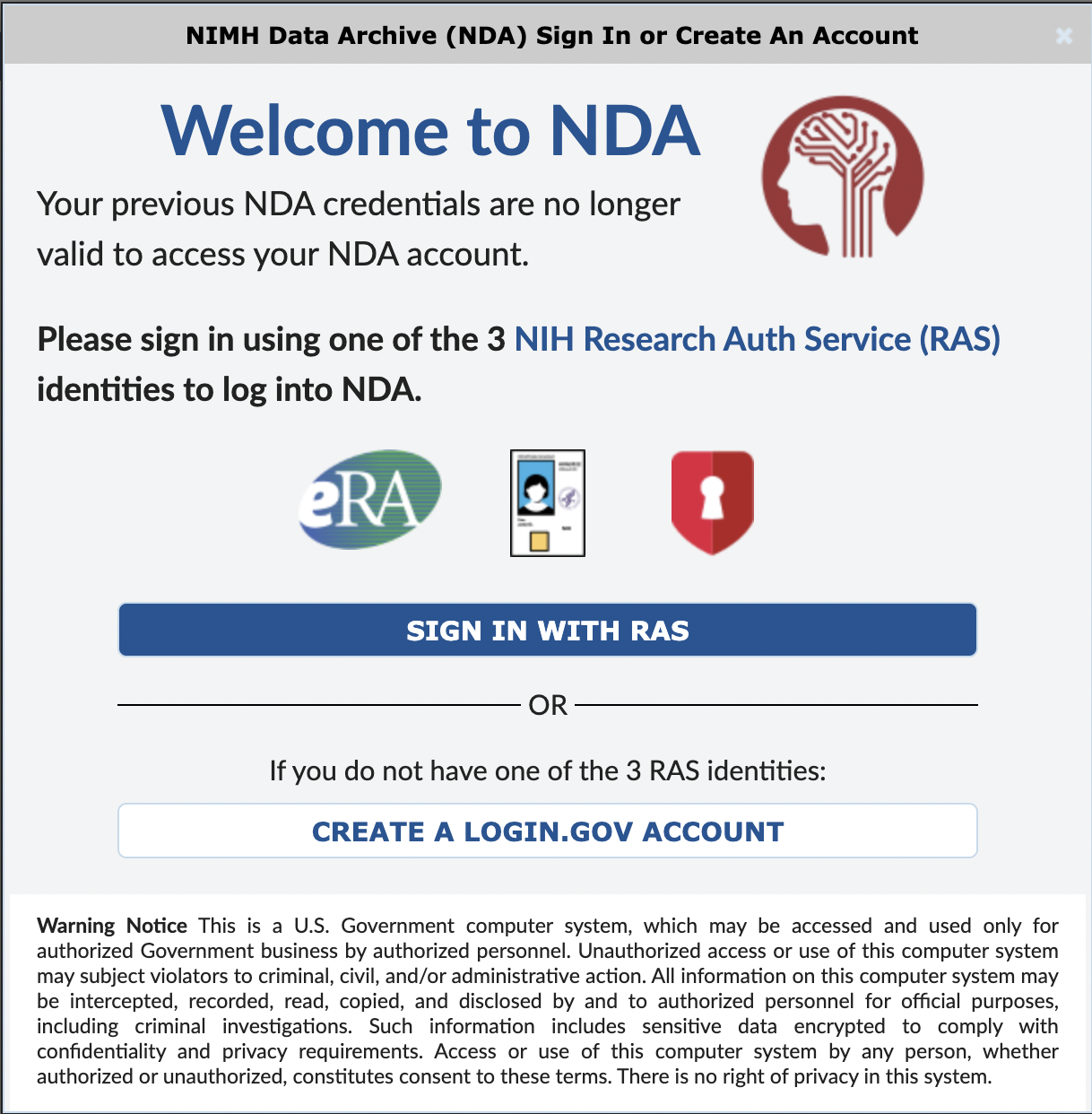
Step 1. Log in to NDA
Log in to your NDA account here.
NDA now uses RAS for login. If you are unable to log in with RAS, please create a Login.gov account associated with your institutional email address. NDA account requests MUST be made using an institutional email address. Account requests made from a personal account will not be honored by NeMO or NDA and will therefore slow down the process of accessing data.
Step 2. Identify Datasets available through the NDA Dashboard
Click on the Data Permissions tab. Scroll down to the NDA Controlled Access Permission Groups. Here you will find all BRAIN/NeMO Data Archives datasets currently available for access request. To the right of your dataset(s) of interest is an 'Actions' dropdown. Select "Request Access". You must work at a research institution that has an active Federal-Wide Assurance in order to initiate a data access request.

Step 3. Data Access Request Tool
This will open the Data Access Request Tool where you will provide information pertaining to your research, institution, and collaborators. Please carefully review the instructions for properly filling out all tabs of the request tool:
A) Request Access Instructions
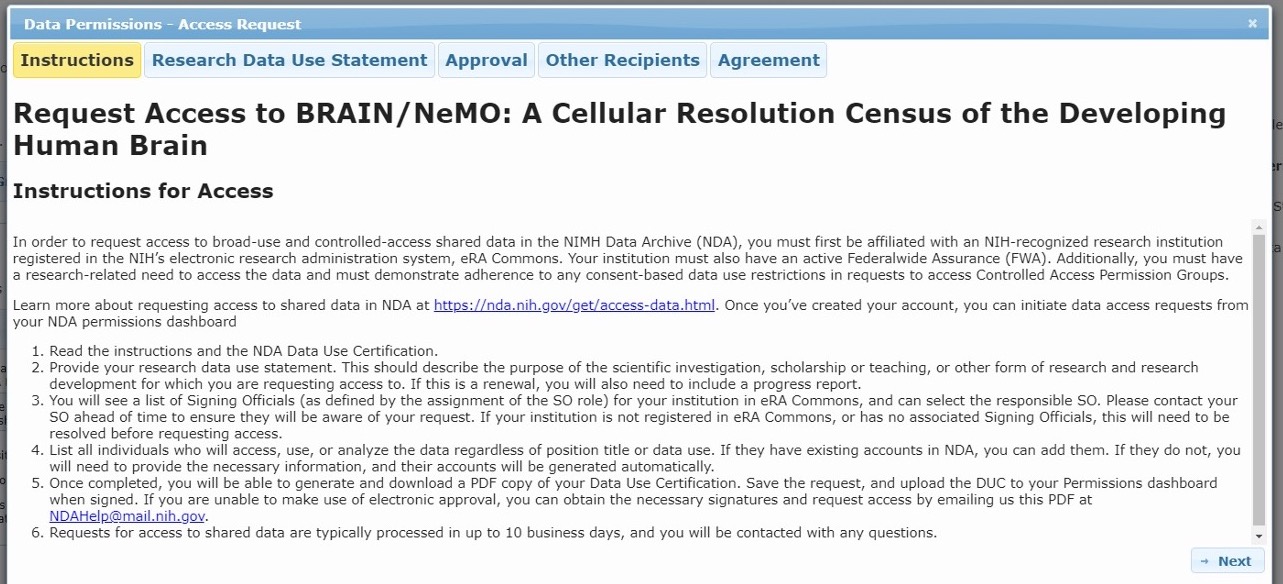
B) Research Data Use Statement
Access requests for controlled access permission groups should include a Research Data Use Statement that appropriately addresses consent-based data use limitations for that permission group. To determine if there are consent-based data use limitations to which authorized researchers must adhere, refer to the “Data Use Limitations” field next to the BRAIN/NeMO dataset of interest in the NDA Controlled Access Permission Group table.
C) Authorized Research Institute
You must work at a research institution that has an active Federal-Wide Assurance in order to initiate a data access request. The signing official(s) associated with your institution will automatically appear as a selectable option.
D) Other Access Recipients
Each data access application is restricted to users from a single institution. If you have collaborators at other organizations, they must submit a separate data access application.
Step 4. Download Data Use Certificate (DUC)
Download the Data Use Certification Agreement PDF from the Agreement tab and complete with signatures of both the investigator and the institutional Signing Official. Contact the NDA Help Desk if you need assistance identifying Signing Officials at your research institution.

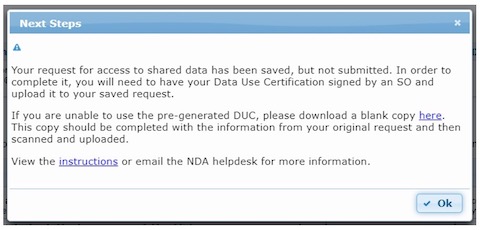
Step 5. Upload signed DUC
Log into the NDA Permissions Dashboard and upload the signed DUC to the “Active Requests” section at the top of the NDA Permissions Dashboard.
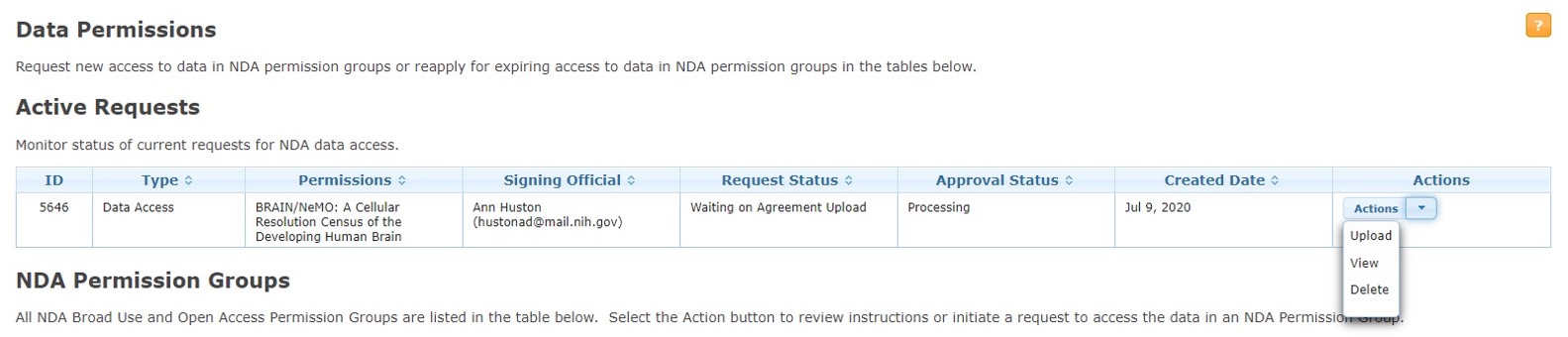
Step 6. Review
Your data access request will be reviewed by an NIH Data Access Committee (DAC). This process typically takes about one week.
Step 7. Access request decision
NDA will inform investigators of a final access decision. At this time, you will need to forward your Access Approval email to the NeMO team at nemo@som.umaryland.edu. Failure to do so will delay our sending you your access credentials. NeMO will grant data access to investigators for one year, after which investigators will need to reapply for access using the process described above.
Restricted access datasets currently available for request at Broad
The Broad Institute's Data Access Committee (DAC) will review data access requests in consideration of the data use limitations on these datasets before approval. For further instructions on how to request and get access to these datasets, go to Requesting access through the Broad's DAC Approval Process.
| Publication Dataset | DUOS Dataset | Description | Data Use Limitations | Dataset Page |
|---|---|---|---|---|
| A concerted neuron-astrocyte program declines in aging and schizophrenia | SCZ_GeneExpression_McCarrollBerretta | We used single-nucleus RNA-seq to analyze 1.2 million nuclei from the prefrontal cortex of 191 persons, including 94 affected by schizophrenia. | Use of this dataset is limited to disease specific research - neurobiologic and psychiatric disorders | dat-bmx7s1t |
| Long somatic DNA-repeat expansion drives neurodegeneration in Huntington's disease | HD_GeneExpression_McCarrollBerretta | To understand how the HTT CAG repeat might underlie pathological changes in Huntington's disease, we developed a method to sequence and measure the CAG repeat together with genome-wide RNA expression in the same cells. This dataset contains long-read PacBio data from the caudate of 9 persons, and single-nucleus RNAseq data from the caudate of 103 persons. | Use of this dataset is limited to disease specific research - neurobiologic and psychiatric disorders | dat-ztfn3cc |
Requesting access through the Broad's DAC Approval Process
To access the data, the requester must first create an account in DUOS using their institutional email address. If the requester's institutional account is not already a Google account, it can be made a Google account by following these instructions. The Signing Official from the requester's institution must register for an institutional Library Card Agreement (“LCA”) in DUOS if one is not already in place. The LCA enables requesters from that institution to request access to data managed by DUOS. The requester will then need to fill out a Data Access Request through DUOS, which will be reviewed by the Broad Institute's Data Access Committee. Once a request is approved, the requester should forward their DUOS approval e-mail to nemo@som.umaryland.edu, and NeMO will authorize access to the data.
Downloading Data
using Google Cloud Platform
If you are a legacy data user and still have access via Aspera, those download instructions can be found here.
Restricted NeMO data is now available through Google Cloud Platform (GCP). There are two mechanisms for access, web interface and command line.
NeMO data is requester pays, therefore downloading by either mechanism requires the use of a Google billing account. In order to avoid incurring large charges for data download, we strongly recommend that you run data analyses on GCP if possible.
Step 1. Creating an institutional Google Cloud account
Once you have received your NDA approval, the next step is to set up an institutional google account. This can be done by going to https://myaccount.google.com/ and selecting Create Account, using the SAME institutional email address that you used for NDA account creation. Not sure what email address is associated with your NDA account? Log in to NDA to access your user dashboard. You will find your email address under your Profile.
For example, if you used the email address janedoe@som.umaryland.edu when you set up your NDA account, then you would now create a new google account using janedoe@som.umaryland.edu
Once set up, notify nemo@som.umaryland.edu as to your institutional google account creation. At this time, we will configure permissions and provide you with the bucket name(s).
Step 2. Setting up GCP Billing
To set up a new billing account go to https://console.cloud.google.com/billing, click CREATE ACCOUNT and follow the instructions.
More information on billing accounts is available here.
Step 3a. Access via the GCP Browser Web Interface
Go to https://console.cloud.google.com/storage/browser/<bucket name without leading gs:// >
For example, https://console.cloud.google.com/storage/browser/human-cortex
In the upper right corner, ensure that you are logged in with your institutional account, not a personal account, or you will not see any data listed.
If it is not already populated, click on the button to select the billing account that you previously created.
Navigate by clicking on the directory listed in the table. Individual files can be downloaded using the GCP Browser. Batch downloads require running the gsutil command line tool. Click on the directory you want to download, and click on DOWNLOAD in the menu directly above the data table. A popup will appear providing the gsutil command to run on your command line. For more on gsutil, read on.
Step 3b. Access via gsutil on the command line
Instructions for installing gsutil as part of the Google Cloud SDK are available here.
To access restricted data, you must authenticate your account. At the command line prompt, type gcloud auth login
Follow the directions on the terminal, which will point you to a URL which you must navigate to from your browser. Here you will log in to your institutional google account. Once logged in, you will be provided with a verification code on your browser screen. Copy this and paste it onto the prompt on the command line. You should then see a message verifying your account, and billing project, if available.
To list bucket contents, gsutil -u [billing-project] ls -l gs://bucket
example:
gsutil -u my-billing-project ls -l gs://human-cortexTo download contents, gsutil -u [billing-project] cp gs://bucket/file.txt /path/to/local/machine/file.txt
example:
gsutil -u my-billing-project cp gs://human-cortex/transcriptome/scell/SSv4/human/raw/Ex_sample_01.fastq.tar /Users/jdoe/Desktop/Ex_sample_01.fastq.tarBatch downloading a directory can be done in the same way, adding the recursive option to the copy command, if necessary, gsutil -u [billing-project] cp -r gs://bucket/* /path/to/local/machine/
example:
gsutil -u my-billing-project cp -r gs://human-cortex/transcriptome/* /Users/jdoe/Desktop