Submitting Data to NeMO - Overview
Data submitted to NeMO falls into three categories:
- Public - data to be immediately distributed openly and freely to the wider research community,
- Embargo - data to be held back, or embargoed, until a specific date, at which point it will be released openly and freely to the wider research community,
- Restricted - Controlled access data to be distributed only to an approved group of users due to consent restrictions, e.g. human data. Often restricted datasets contain a combination of private (raw reads, alignments) and public (counts, peaks) datatypes. In such instances, users will submit all data to the Restricted area and we will work with them to obtain consent for releasing any files publicly.
In this document:
- Requesting a NeMO Aspera Account
- Submitting a File Manifest
- Submitting Data
- Processing Workflow
- Restricted Data
- Tracking Submissions
- Reporting to NIH
- Leaving a lab
Requesting a NeMO Aspera Account
New data submitters can register for an account at nemoarchive.org/register. Upon doing so, please send an email to nemo@som.umaryland.edu with the following information:
-
requested username
-
grant under which data was generated
-
PI of lab in which data was generated
-
whether data is public, for embargo or restricted*
-
NEW: Due to new security policies, NeMO Archive will only approve submission account requests from institutional email addresses. Personal email addresses (e.g. @gmail.com, @outlook.com etc) will not be accepted
*Restricted data submitters are asked to request account usernames with an appended "-restricted", e.g. doe-restricted. If you will be submitting both public and controlled access data, you will need to create two accounts, one without and one with the "-restricted" tag. Similarly, if you are submitting to multiple restricted projects (e.g. BICAN and SCORCH) we ask that you create a unique user for each project.
Someone from the NeMO team will follow up with you regarding any other questions about your data or the submission process.
Prior to leaving a lab or otherwise changing roles in a way that would alter your access to data avaliable at NeMO, please complete the offboarding form here.
Submitting a File Manifest
All submissions to NeMO Archives, whether public or private, begin with upload of a file manifest including MD5 checksums through the User Dashboard available to logged in users at https://nemoarchive.org/login or via an API. More instructions on submission are below. This enables us to collect some basic information in order to organize and process your data properly for release.
Detection and validation of a properly formatted manifest will trigger an email message providing the Aspera submission path for your data, see below.
Manifest format
For SCORCH submitters, please see the SCORCH specific documentation located here (link).
For other submitters:
-
Excel-friendly manifest file with field descriptions and controlled vocabularies - Please contact NeMO to discuss addition of new terms to any of the controlled vocabularies.
All manifest files must contain the following fields. Do not delete headers for unused, optional columns:
- File name - must be unique, described below
- Sample ID
- Program (e.g. BICCN)
- Sub-program (e.g. U19 Zeng)
- Lab
- Species
- Modality
- Technique
- Subspecimen type
- Data type
- File type
- Access
- Checksum - described below
- Anatomical site
- Counts data only fields (optional):
- Counts pipeline
- Aligned data only fields (optional):
- Read aligner
- Genome build
- Gene set release
- BRAIN initiative only fields
- BCDC Project name
- BCDC data collection
- Controlled access only fields (optional):
- Usage (e.g. Limited/GRU/NA)
- Institutional Certification ID
- Donor id/name
- Tissue provider
The manifest file must contain one row, including an MD5 checksum, for every file included in the submission. If you're submitting a tarball, the manifest should contain one row for every file within the tarball. For submission of multiple, or chunked, tarballs, there is no need for a manifest per tarball, all component files can be included in a single manifest. Individual manifests are limited to 25,000 files. If you are submitting more than 25,000 files, please seperate into multiple manifests.
The first column should contain the file name only, with no path information. File name must be unique. In the case of sample-specific bundles, for example mex bundles containing a genes, features and barcodes file, prepend the sample id to your file name. We will strip that off during processing and place the three files in a sample id directory, as expected by many downstream tools. As an example, three files are submitted to NeMO with the following sample names:
sample1_genes.tsv
sample1_features.tsv
sample1_barcodes.tsvThese files will be released as three files within the sample1 subdirectory:
sample1/genes.tsv
sample1/features.tsv
sample1/barcodes.tsvImportant:
Save your manifest as a tab seperated file. The manifest file can have any prefix, however the base filename must be manifest.tsv.
Manifest submission and validation
To submit your manifest for validation, log into NeMO using your NeMO Aspera credentials. Once logged in, select Dashboard in the top navigation. This will take you to a page with links to make a submission, or view your current submissions (described below).
Upon upload, file manifests will be immediately validated for:
- complete header row
- presence of checksums and all other required fields
- proper controlled vocabularies
- detection of unexpected file extensions
- presence of all components of various bundle types
Validation failure in any row(s) will result in an error message displayed on the submission page, accompanied by an invalid manifest validation email with additional details about the error, if applicable. Once you have corrected the errors, your updated validation file is submitted in the same way and undergoes the same validation.
Validation success will result in notification, typically within minutes, via email, of the Aspera path to which you will submit your data, as described below. Be sure to submit to the provided path, as any other submissions will be ignored and routinely deleted.
NEW: To help users validate their manifest, we have implemented a 'dry-run' feature which allows users to test if their manifest meets the validation criteria without recording each attempt in our system. While using the 'dry-run' feature (i.e. selecting the 'dry-run' box on the manifest upload page) submitters are notified of the results of validation via email similar to a normal submission but no further action is taken and the submission is NOT listed in the user's submission dashboard.
To perform a dry run using the submission API, add "dryrun=y" to the submission creation endpoint as a query string parameter. More information can be found here Dry runs using submission API
Submitting Data
NeMO accepts both raw and derived data types of the file extensions listed here. If you plan to submit data extensions not currently on this list, please let us know ahead of time to avoid manifest upload and/or processing delays, by emailing nemo@som.umaryland.edu. NeMO Archive discourages the submission of aggregate files through our ingest process (i.e. files containing counts from multiple samples). If you have a need to submit aggregated files, please contact us prior to attempting a submission.
Depending on the size of your submission, it may be preferable to submit data files individually or within a tarball. Either is fine. We don't pre-define any specific directory structure for your submission, however we ask that you do not submit tarballs within a tarball, as that will delay processing. Files will be reorganized during ingest based on metadata provided in the manifest.
Uploading using Aspera
In order to submit data to NeMO, you will first need to download and install the IBM Aspera Command Line Interface, available from the IBM website. You will find IBM Aspera CLI under Developer resources. Installation instructions are available here.
Once installed, the ascp utility will be available for use at the command line. Have your NeMO Aspera credentials handy, as commands to initiate an upload will result in a prompt for your password.
Uploading with the Aspera client uses the following syntax:
$ ascp [-l <Maximum download speed>] [-k 2] [-m <Minimum download speed>] [-Q] [-T] \
<path to local file or directory> <username>@aspera.nemoarchive.org \
provided/path/to/NeMO/directory/All parameters between the [ ... ] brackets are optional parameters described here:
l - Allows for the user to set a maximum upload/download speed that aspera should attempt to stay at or below for the duration of the transfer. A speed in Megabits must be provided with this flag.
k - Allow for resumable data transmission in case an interruption occurs.
m - Allows for the user to set a minimum upload/download sped that aspera should attempt to stay at or above for the duration of the transfer. A speed in Megabits must be provided with this flag.
Q - Turns adaptive rate on. Adaptive rate controls the speed of aspera with a goal of not dominating the bandwidth available. Very useful on busy networks that may have other transfers ongoing.
T - Turns encryption off. Turning encryption off will allow for a maximum throughput transfer but should not be used if data being uploaded is sensitive.In the following example, my_data.tar.gz is being transferred to NeMO,
$ ascp -l 1000M -k 2 -QT /home/user/my_data/my_data.tar.gz user123@aspera.nemoarchive.org:v34kltf7/where v34kltf7 is the directory name that was provided to user123 upon manifest validation. Uploading a directory of files would not require any changes as Aspera will recognize that a folder is being transferred and will recursively step through ensuring all files found in the directory are transferred.
$ ascp -l 1000M -k 2 -QT /home/user/my_data/ user123@aspera.nemoarchive.org:v34kltf7/It is important to note that the above does NOT guarantee that the transfer will proceed at 1000Mbps. This is only a target rate. The NeMO Aspera server is capped to 1000Mbps (1Gbps) in aggregate, so the bandwidth is shared across all concurrent users. This may cause some contention, resulting in a lower effective transfer speed, depending on the demand.
It is not possible to delete or update data using the Aspera CLI, however submitting a file of the same name will overwrite the previous file. Should you wish to delete data, reach out to a NeMO team member at nemo@som.umaryland.edu.
Important:
Once your data upload has completed, please run one more Aspera upload of a single, empty file named 'DONE' ,with no extension, in the top level of your submission directory (i.e. not within a folder). This will prompt our system to begin scanning your submission, if the DONE file is uploaded before other file uploads are complete, the submission will fail.
Processing Workflow
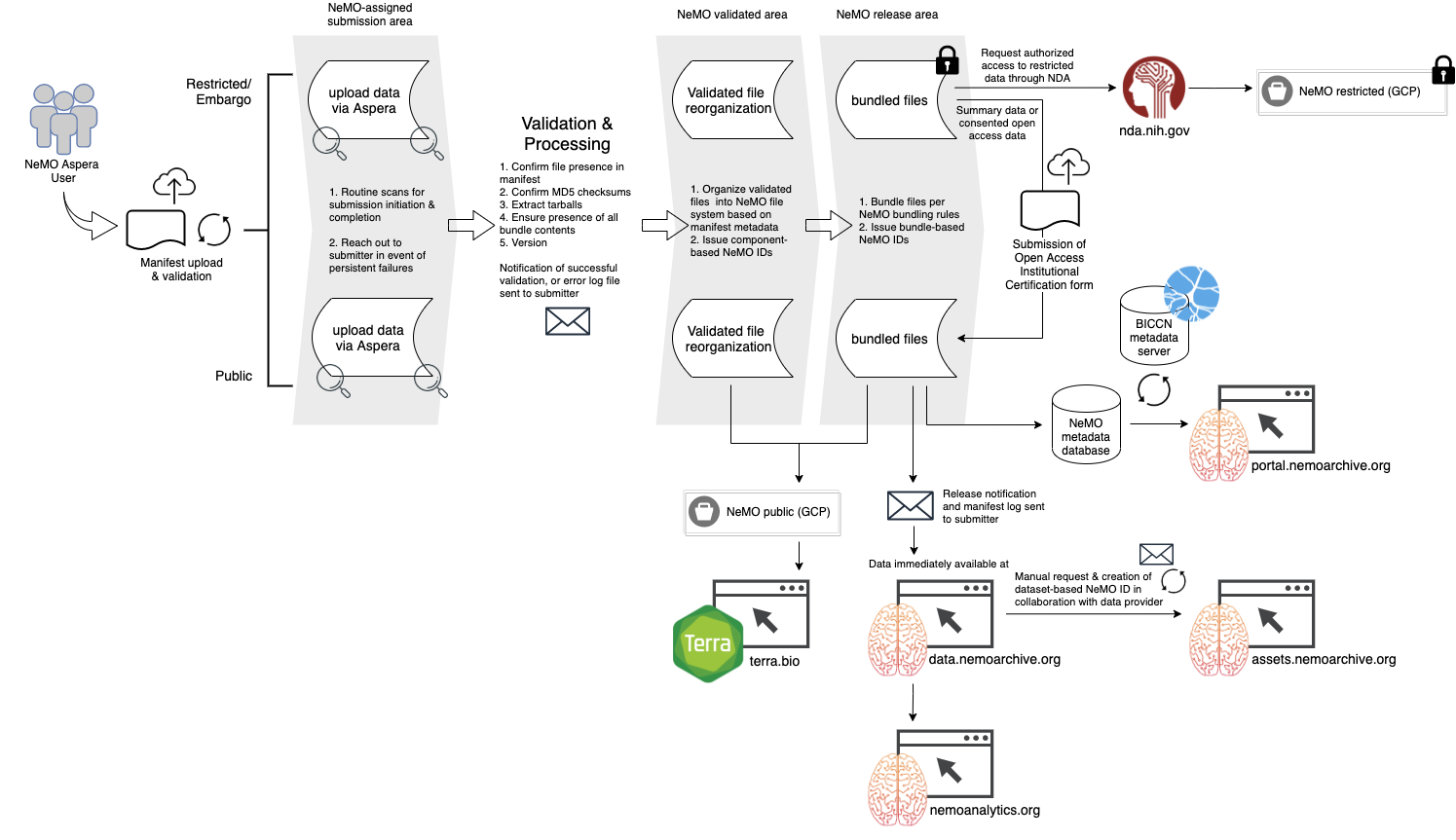
Upon successful bundling of data to the release area (either public or restricted), submitters will receive a "Successful ingest" email containing your validated manifest with additional fields capturing final file location and NeMO identifiers for individual files. This email will also list any directories or files skipped due to not being listed in the manifest, for your review.
Restricted Data
For restricted, or controlled access, data submissions, the validation and ingest process is the same.
As described above, NeMO Aspera accounts for restricted data submissions should be created with an appended "-restricted", e.g. doe-restricted. Restricted accounts need to be set to mount to the proper area on our restricted server. A NeMO team member will reach out, distinctly from the automated account approval email, once your account is ready for restricted data submission, so please plan accordingly for this minimal extra time required prior to data submission.
Restricted Data Release documentation outlines steps required to make restricted datasets available to approved users or the public, as applicable.
Tracking Submissions
Submitters can track the progress of data submissions from manifest validation through release to the NeMO Portal. To track submissions, log into the NeMO website using your NeMO Aspera credentials. Once logged in, select Dashboard in the top navigation, and select Submission status.
This image shows the status for an individual manifest. Clicking on any of the completed, green checkmarks provides additional details: date, time and explanation of the completed step.
Incomplete steps are shown in gray, failed steps in red. Failed steps following the QC step will be resolved internally by the NeMO team, and we will reach out if you need to resubmit all or a portion of your data.
If you are a project manager, a PI or other role responsible for tracking submissions, and would like to have access to the tracking dashboard for one or a number of labs or submitters, please create a NeMO Aspera account at nemoarchive.org/register and email nemo@som.umaryland.edu explaining your role and tracking request.
Reporting to NIH
Depending on funding source, data submitters may be required to submit quarterly proof of data submissions. At this time, BICCN is accepting the "Successful ingest" email received once data is successfully bundled as a submission receipt for an individual manifest.
Leaving a lab or position & verification of continuing accounts
In addition to regular audits of lab members by NeMO staff (individuals noted as PIs will regularly be asked to verify the status of members of their lab) individuals leaving a lab or otherwise changing roles in a way that would alter your access to data avaliable at NeMO, should complete the offboarding form here.